VAMPhyRE: Virtual Analysis Method for Phylogenomic fingerPrint Estimation.
- What is VAMPhyRE?
- Virtual Hybridization
- Estimating probe size
- Other Virtual Hybridization parameters
- Virtual Genomic Fingerprints (VGFs)
1. What is VAMPhyRE?
The Virtual Analysis Method for Phylogenomic fingeRprint Estimation
(VAMPhyRE) is a bioinformatics technique aimed for whole-genome
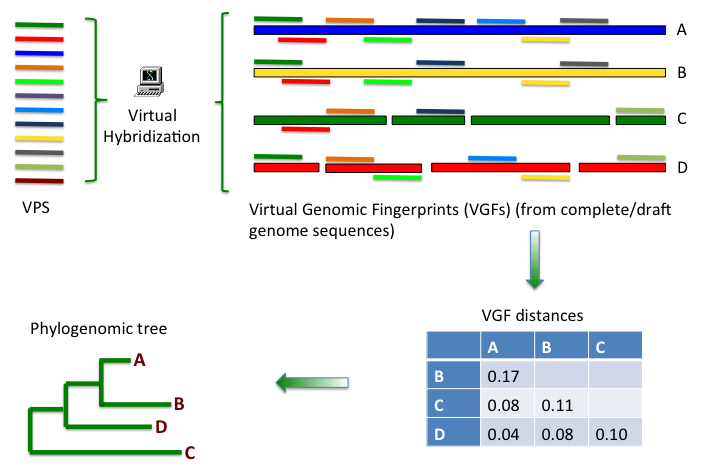
comparisons and phylogenomic analysis using Virtual Genomic
Fingerprints (VGFs). VGFs are calculated by Virtual Hybridization (VH),
which is a computational method that searches potential hybridization
sites for short probes on genomic sequences. The collection of
potential hybridization sites in a target genome constitutes a
hybridization pattern called Virtual Genomic Fingerprint (VGF).
VGFs can be used to estimate genome similarity between pairs of
genomes. Pairwise genome distances are calculated from the number of
homologous sites shared between the VGFs of two genomes to be
compared. Then, a table of distances between pairs of genomes can
be used for calculating phylogenomic trees (figure 1).
2. Virtual Hybridization
The Virtual Hybridization analysis (VH) is a computational approach that searches potential hybridization sites for short probes on genomic sequences. This search locates potential hybridization sites between genome sequences and the probes in a defined set (The VAMPhyRE probe set), based on the number of complementary bases allowing a defined number of mismatches. Additionally, the program can perform a thermodynamic analysis for predicting the thermal stability of the duplexes formed between probes and potential hybridization sites, which can be used for the development of microarray devices.
3. Estimating Probe Size
Users can select a predesigned set of probes with lengths ranging from 8 to 13-mer. Each VAMPhyRE Probe Set (VPS) has been selected through a process aimed to maximize the differences between the probes in the set. This property will allowing explore efficiently the genome sequence complexity. The probe size (k) must be estimated from the genome length (L). The next formula can be used to an approximate estimation of probe sizes as a function of genome size:
k = (1/2) * Log2 (4L)
The table resumes the properties of the probe sets that are available on the server.
| Probe set | Number | Targets |
| VPS8 | 342 | genes, viruses |
| VPS9 | 1,327 | viruses |
| VPS10 | 4,936 | viruses |
| VPS11 | 20,951 | viruses, small bacterium |
| VPS12 | 1,327 | viruses, bacterium |
| VPS13 | 15,265 |
bacterium |
4. Other Virtual Hybridization Parameters
Target sequences. Target sequences must be provided as a FASTA file for multiple sequences. Sequences are assumed that are not aligned.
Internally the title of each sequence is edited and truncated to 10 characters, therefore we encourage the users to check the titles to avoid repeated ones, once they are truncated.
The maximum size of the files is 15 MB. Be aware that with slow connections the upload process can take a long time.
Free energy threshold. Please set this value to zero as default.
Free energy values are not necessary for genome comparisons.
However, the free energy can be useful for simulation of microarray experiments.
Free energy threshold can be used to filter hybridization sites according to their stability.
Typically the free energy threshold can be set between -15 and 0 Kcal/mol for UFC probes.
Allowed number of mismatches. If mismatches are allowed the number of potential hybridization sites increases.
By allowing a single mismatch, and using the recommended probe size calculated by the formula (1) the virtual hybridization explores a higher proportion of the genome,
which is convenient for genome comparisons. The recommended number of mismatches is 1.
Strand. Virtual hybridization searches probe similarities with the direct or the complementary strand.
Hybridization occurs then with the opposite strand. Moreover, searches can be done with both strands simultaneously.
Show hybridization sites. Virtual Hybridization analysis can show all the hybridization sites with less or at most the allowed number of mismatches (which are defined in the main parameters).
Alternatively, the results can be filtered to show only those sites with the exact number of mismatches.
Additionally, the results can be adjusted to show all the hybridization sites with less or at most the allowed number of mismatches, as well as sites with more mismatches but with thermal stability inside of the range of the previous results.
The last option can be useful when using the VH analysis to interpret experimental results, where the user may be interested in listing all the possible hybridizations sites inside a range of stability.
Ambiguous hybridization sites. By default, VH analysis will list all the hybridization sites of a particular probe with a given genome.
Alternatively, the search can be configured to show only the most stable site.
Format of the VH results. This is an internal-staff parameter used to configure the results for their use in a new or previous version of the software.
Please use the New format.
Calculate global table. A global table resumes the results of the search.
This table will list all the probes and all the genomes and will detail if the probe can hybridize with a given genome.
This table can be easily imported to spreadsheet programs like Excel or LibreOffice. This file can be used in the program characters (included in this site) for comparing genome fingerprints.
Report of sites in the table The global table can report if each probe can (1) or cannot (0) hybridize with each genome. This report can be configured also to show the number of times that each probe hybridizes with each genome.
Alternatively, the report can show the total number of bases that can hybridize considering all the hybridization sites.
5. Virtual Genomic Fingerprints (VGFs)
The result of the virtual hybridization analysis is a list of the probes that can hybridize with a given genome under the conditions previously defined. This list includes detailed information about the hybridization positions, strand, targeted sequence (sequence of the site of hybridization), and free energy for the duplex probe/hybridization-site. This list constitutes a Virtual Genomic Fingerprint (VGF) (see figure 2).

